ongoing project
Stain Normalization
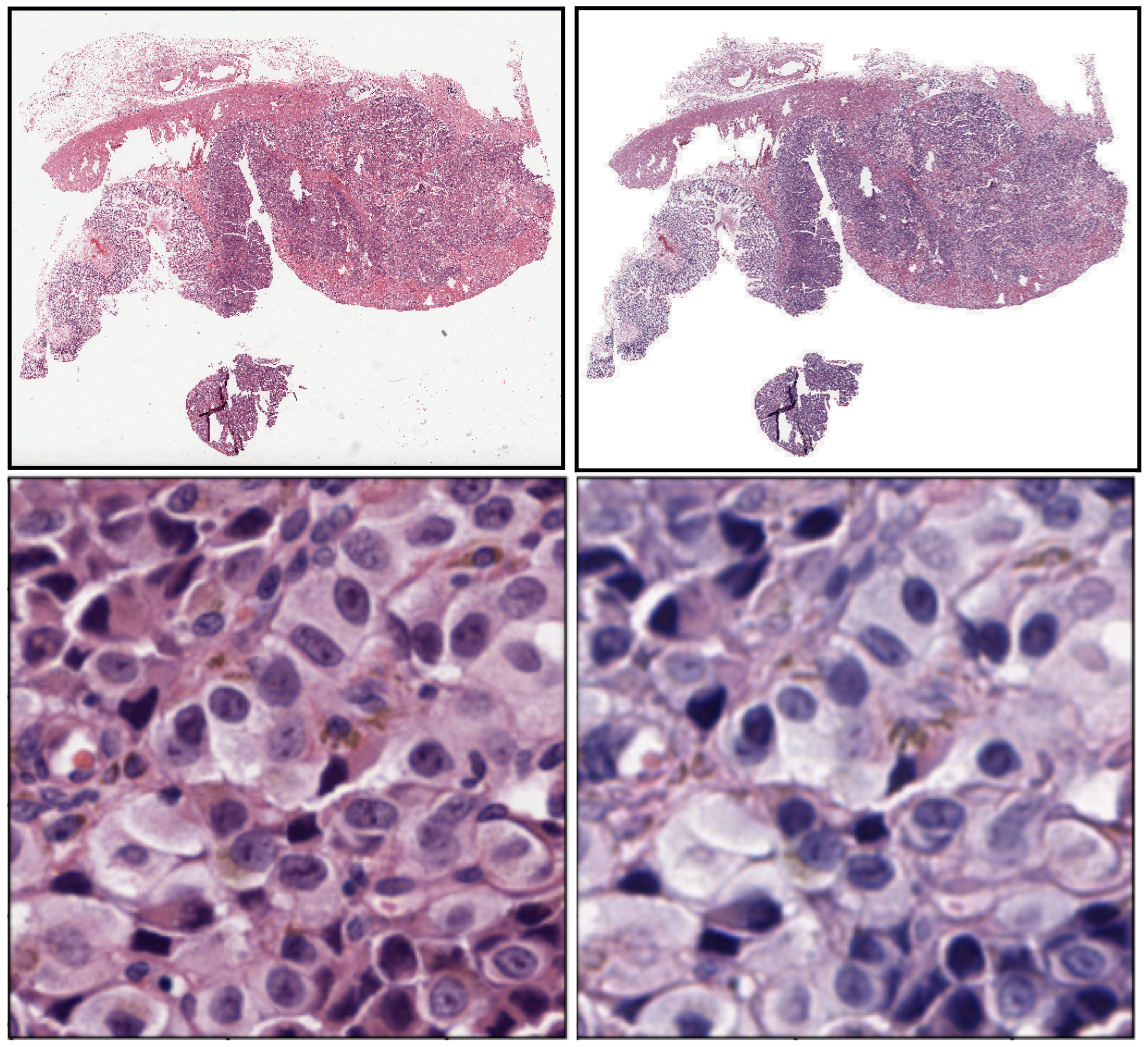
Scientific Premise:
Inconsistencies in the preparation of histology slides make it difficult to perform quantitative analysis on tissue images, including the training of deep learning models. We aim to perform an in-depth characterization of these inconsistencies to make pragmatic recommendations.
Motivation:
To develop deep learning algorithms for histopathology, it is essential to have access to large amounts of data in order to develop efficient and accurate pretrained models. There already exists a collection of publicly available pathological data published by the NIH (TCGA dataset) with over 20,000 images representing all tissue types. We would like to utilize this data in order to pretrain our models. The origins of the TCGA dataset encompasses hundreds of institutions from across the United States. This results in the pathological image data having highly variable staining methods and histological preparation. For the training of the deep learning models, it is necessary that the data has a standardized stain for the deep learning model to reduce the batch effect, which can degrade model performance. Preferably, the TCGA staining should match the institution where the models will be implemented. In this case, we utilized histology slides at Dartmouth Hitchcock Medical Center in Lebanon, NH as a reference.
The scope of our Stain Normalization Project is to evaluate various computational histological stain normalization techniques and different failure modes. These techniques will incorporate both Deep Learning and non-deep learning approaches. As a deliverable, we will identify effective stain normalization techniques.