ongoing project
Multimodal Modeling
Scientific Premise:
That the synthesis of information across multiple biomedical data modalities can provide a more holistic and accurate depiction of the patient’s health.
Motivation:
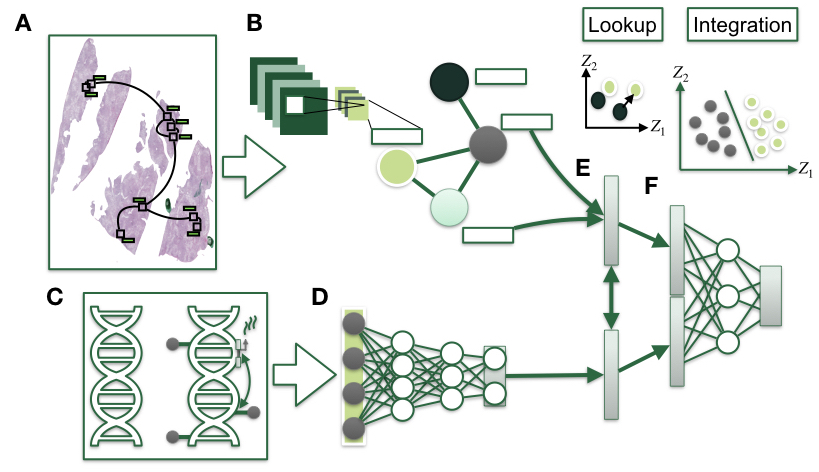
Genetic and epigenetic alterations drive oncogenesis, which may be reflected by morphological changes at the cellular level observable with histopathological approaches to diagnosis and staging. The underlying data modalities, such as DNA methylation (DNAm), gene expression, copy number variation (CNV), SNPs, and histopathological imaging, each by themselves present unique disease-specific and prognostic information that when combined could potentially improve the diagnostic and prognostic performance. However, these data are highly complex and heterogenous, high dimensional, require large amounts of storage, and are costly to procure. In addition, machine learning models that integrate multimodal information must grapple with the fact that the memory required to process just a single training instance can exceed 50 GB. All of these factors demonstrate the nontriviality of these modality representations and explain why relatively little research has integrated histopathology images and genomics modalities. Deep learning approaches that heuristically project data into nonlinear manifolds while taking into account interactions between the features have achieved state-of-the-art performance for analyzing histology slides and other omics modalities individually. These approaches create compressed representations of data but do not explicitly model prognostic information that is shared and disjoint, cannot scale to the available computational resources and remain unvalidated and thus do not present viable multimodal solutions that can be readily deployed into the clinical workflow.
Developments in deriving succinct representations of information across WSI and high-dimensional omics data now provide an avenue from which to train models that may utilize both media for prediction tasks. Existing methods have demonstrated that such integration is possible. Our group aspires to build on these methods by leveraging the topological properties of both omics and whole slide imaging and modeling shared and dissimilar latent profile across two or more modalities to improve and standardize disease diagnosis and prognostication.
While key somatic alterations may drive initiating oncogenic changes, corresponding changes in morphology may be utilized to improve the performance of multimodal approaches or contribute to a better understanding of disease pathology. Developments in deriving compact representations of information across WSI and high-dimensional omics data via self-supervised and weakly-supervised approaches have demonstrated promising results for multimodal prognostic prediction models. We are working on methods that pay careful attention to the representation method for the WSI via graphs, incorporate biological knowledge by grouping CpGs by colocalized genomic contexts and more directly incorporate mutual information for multiple modalities within the objective function.
We aim to further develop promising DNAm and WSI data representation techniques for eventual incorporation into multimodal deep learning biomedical technologies that can generalize to modalities outside of DNAm and WSI, interoperable between modalities from radiomics, to natural language processing, gene expression, amongst others. The aim for subcomponents of these approaches is to bridge gaps in oncology care in the global health setting by improving the compact representation of patient’s health profile by learning from multiple modalities. These approaches may improve predictive accuracy in data scarce regions, especially where the quality of data from one modality may be lacking or of particularly low quality.
Manuscripts:
- Levy, J. Emerging Diagnostic and Investigative Technologies: Validation of Deep Learning Technologies for Dna Methylation and Histopathology. (Dartmouth College, 2021).
- Azher, Z. L., Vaickus, L. J., Salas, L. A., et al. Development of Biologically Interpretable Multimodal Deep Learning Model for Cancer Prognosis Prediction. 2021.10.30.466610 https://www.biorxiv.org/content/10.1101/2021.10.30.466610v1 (2021) doi:10.1101/2021.10.30.466610.
- Haudenschild, C., Vaickus, L. & Levy, J. Configuring a federated network of real-world patient health data for multimodal deep learning prediction of health outcomes. 2021.10.30.466612 https://www.biorxiv.org/content/10.1101/2021.10.30.466612v1 (2021) doi:10.1101/2021.10.30.466612.
- Levy, J., Haudenschild, C., Barwick, C., et al. Topological Feature Extraction and Visualization of Whole Slide Images using Graph Neural Networks. Pac Symp Biocomput 26, 285–296 (2021).