Welcome to PathFlowAI’s documentation!¶
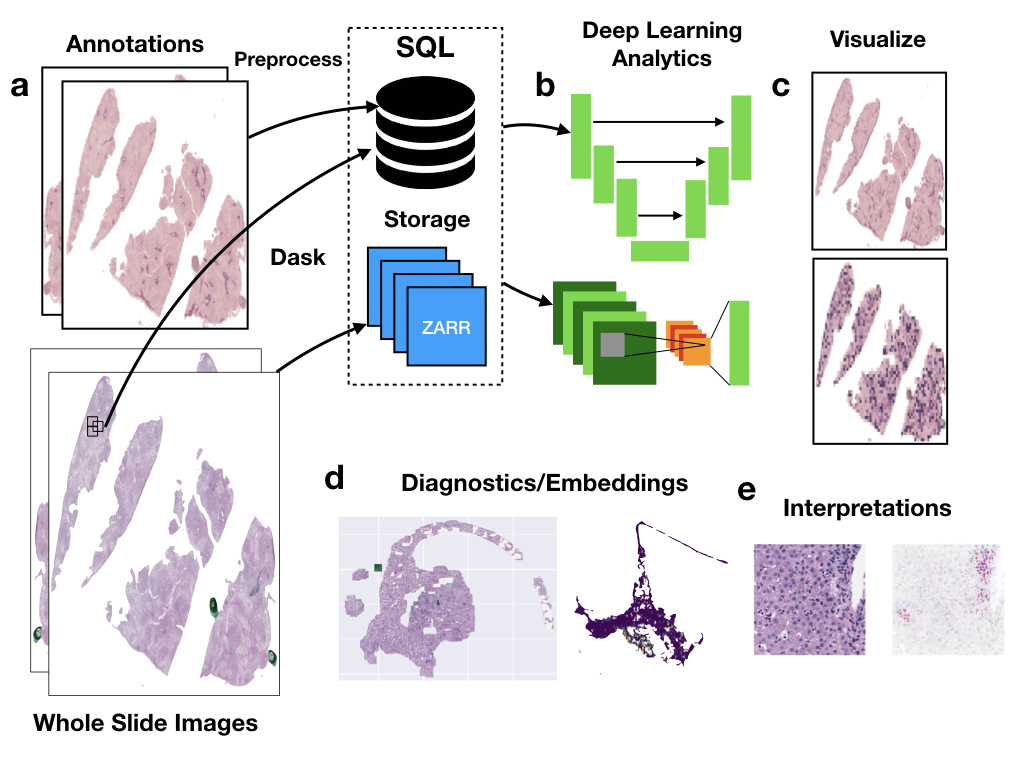
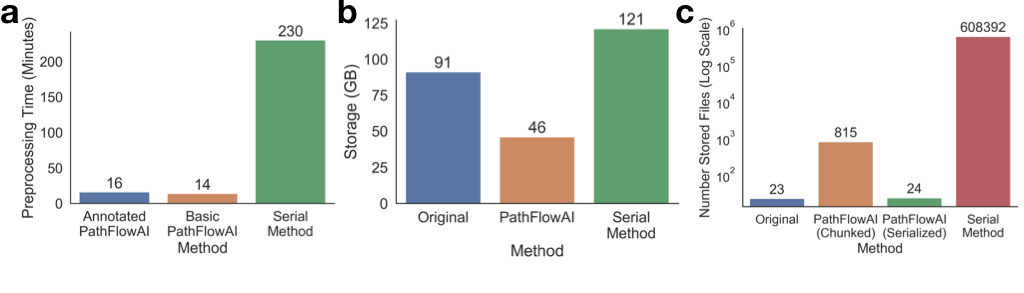
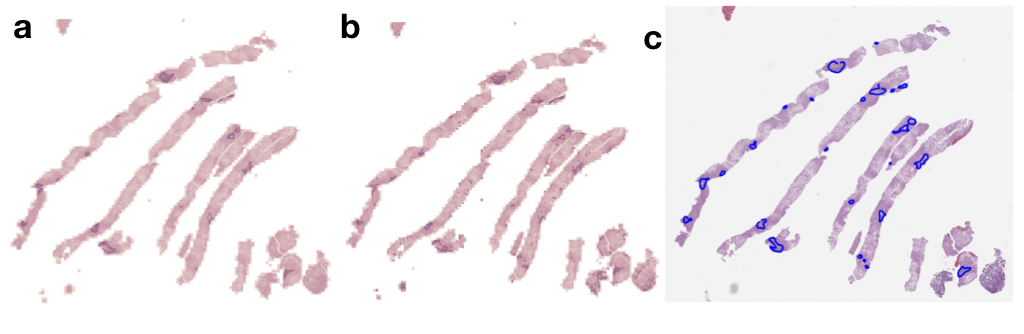
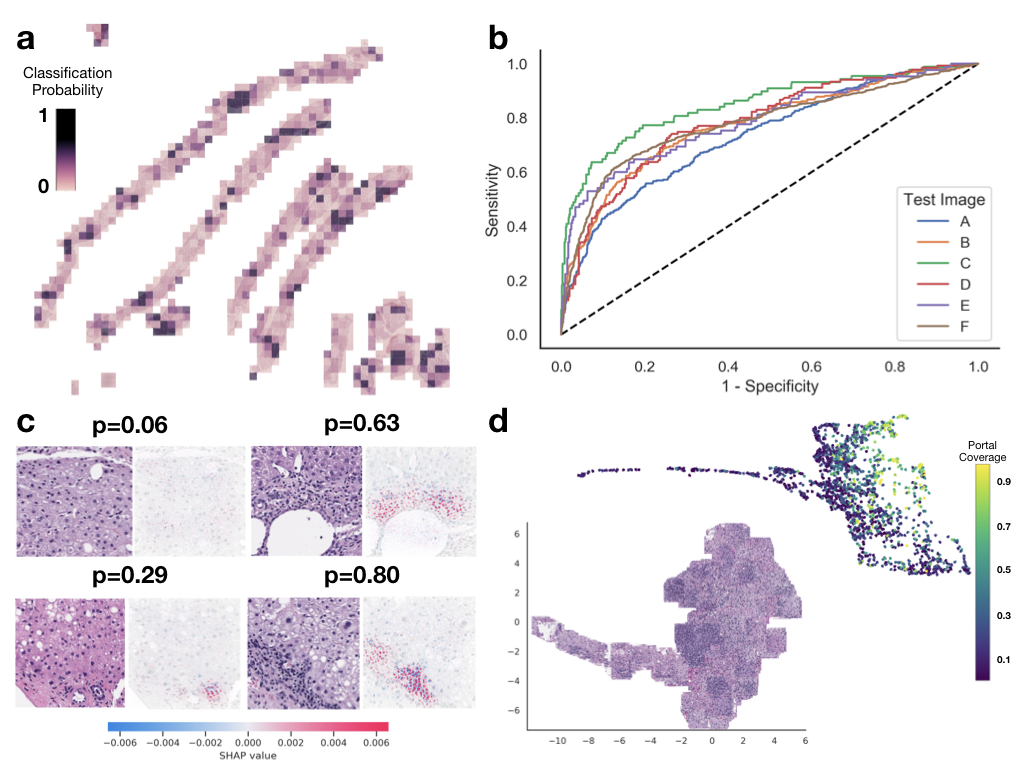
pathflowai-preprocess¶
pathflowai-preprocess [OPTIONS] COMMAND [ARGS]...
Options
-
--version¶ Show the version and exit.
alter_masks¶
Map list of values to other values in mask.
pathflowai-preprocess alter_masks [OPTIONS]
Options
-
-i,--mask_dir<mask_dir>¶ Input directory for masks. [default: ./inputs/]
-
-o,--output_dir<output_dir>¶ Output directory for new masks. [default: ./outputs/]
-
-fr,--from_annotations<from_annotations>¶ Annotations to switch from. [default: ]
-
-to,--to_annotations<to_annotations>¶ Annotations to switch to. [default: ]
collapse_annotations¶
Adds annotation classes areas to other annotation classes in SQL DB when getting rid of some annotation classes.
pathflowai-preprocess collapse_annotations [OPTIONS]
Options
-
-i,--input_patch_db<input_patch_db>¶ Input db. [default: patch_info_input.db]
-
-o,--output_patch_db<output_patch_db>¶ Output db. [default: patch_info_output.db]
-
-fr,--from_annotations<from_annotations>¶ Annotations to switch from. [default: ]
-
-to,--to_annotations<to_annotations>¶ Annotations to switch to. [default: ]
-
-ps,--patch_size<patch_size>¶ Patch size. [default: 224]
-
-rb,--remove_background_annotation<remove_background_annotation>¶ If selected, removes 100% background patches based on this annotation. [default: ]
-
-ma,--max_background_area<max_background_area>¶ Max background area before exclusion. [default: 0.05]
preprocess_pipeline¶
Preprocessing pipeline that accomplishes 3 things. 1: storage into ZARR format, 2: optional mask adjustment, 3: storage of patch-level information into SQL DB
pathflowai-preprocess preprocess_pipeline [OPTIONS]
Options
-
-npy,--img2npy¶ Image to numpy for faster read. [default: False]
-
-b,--basename<basename>¶ Basename of patches. [default: A01]
-
-i,--input_dir<input_dir>¶ Input directory for patches. [default: ./inputs/]
-
-a,--annotations<annotations>¶ Annotations in image in order. [default: ]
-
-pr,--preprocess¶ Run preprocessing pipeline. [default: False]
-
-pa,--patches¶ Add patches to SQL. [default: False]
-
-t,--threshold<threshold>¶ Threshold to remove non-purple slides. [default: 0.05]
-
-ps,--patch_size<patch_size>¶ Patch size. [default: 224]
-
-it,--intensity_threshold<intensity_threshold>¶ Intensity threshold to rate a pixel as non-white. [default: 100.0]
-
-g,--generate_finetune_segmentation¶ Generate patches for one segmentation mask class for targeted finetuning. [default: False]
-
-tc,--target_segmentation_class<target_segmentation_class>¶ Segmentation Class to finetune on, output patches to another db. [default: 0]
-
-tt,--target_threshold<target_threshold>¶ Threshold to include target for segmentation if saving one class. [default: 0.0]
-
-odb,--out_db<out_db>¶ Output patch database. [default: ./patch_info.db]
-
-am,--adjust_mask¶ Remove additional background regions from annotation mask. [default: False]
-
-nn,--n_neighbors<n_neighbors>¶ If adjusting mask, number of neighbors connectivity to remove. [default: 5]
-
-bp,--basic_preprocess¶ Basic preprocessing pipeline, annotation areas are not saved. Used for benchmarking tool against comparable pipelines [default: False]
remove_basename_from_db¶
Removes basename/ID from SQL DB.
pathflowai-preprocess remove_basename_from_db [OPTIONS]
Options
-
-i,--input_patch_db<input_patch_db>¶ Input db. [default: patch_info_input.db]
-
-o,--output_patch_db<output_patch_db>¶ Output db. [default: patch_info_output.db]
-
-b,--basename<basename>¶ Basename. [default: A01]
-
-ps,--patch_size<patch_size>¶ Patch size. [default: 224]
pathflowai-visualize¶
pathflowai-visualize [OPTIONS] COMMAND [ARGS]...
Options
-
--version¶ Show the version and exit.
extract_patch¶
Extract image of patch of any size/location and output to image file
pathflowai-visualize extract_patch [OPTIONS]
Options
-
-i,--input_dir<input_dir>¶ Input directory for patches. [default: ./inputs/]
-
-b,--basename<basename>¶ Basename of patches. [default: A01]
-
-p,--patch_info_file<patch_info_file>¶ Datbase containing all patches [default: patch_info.db]
-
-ps,--patch_size<patch_size>¶ Patch size. [default: 224]
-
-x,--x<x>¶ X Coordinate of patch. [default: 0]
-
-y,--y<y>¶ Y coordinate of patch. [default: 0]
-
-o,--outputfname<outputfname>¶ Output extracted image. [default: ./output_image.png]
-
-s,--segmentation¶ Plot segmentations. [default: False]
-
-sc,--n_segmentation_classes<n_segmentation_classes>¶ Number segmentation classes [default: 4]
-
-c,--custom_segmentation<custom_segmentation>¶ Add custom segmentation map from prediction, in npy [default: ]
overlay_new_annotations¶
Custom annotations, in format [Point: x, y, Point: x, y … ] one line like this per polygon, overlap these polygons on top of WSI.
pathflowai-visualize overlay_new_annotations [OPTIONS]
Options
-
-i,--img_file<img_file>¶ Input image. [default: image.txt]
-
-a,--annotation_txt<annotation_txt>¶ Column of annotations [default: annotation.txt]
-
-ocf,--original_compression_factor<original_compression_factor>¶ How much compress image. [default: 1.0]
-
-cf,--compression_factor<compression_factor>¶ How much compress image. [default: 3.0]
-
-o,--outputfilename<outputfilename>¶ Output extracted image. [default: ./output_image.png]
plot_embeddings¶
Perform UMAP embeddings of patches and plot using plotly.
pathflowai-visualize plot_embeddings [OPTIONS]
Options
-
-i,--embeddings_file<embeddings_file>¶ Embeddings. [default: predictions/embeddings.pkl]
-
-o,--plotly_output_file<plotly_output_file>¶ Plotly output file. [default: predictions/embeddings.html]
-
-a,--annotations<annotations>¶ Multiple annotations to color image. [default: ]
-
-rb,--remove_background_annotation<remove_background_annotation>¶ If selected, removes 100% background patches based on this annotation. [default: ]
-
-ma,--max_background_area<max_background_area>¶ Max background area before exclusion. [default: 0.05]
-
-b,--basename<basename>¶ Basename of patches. [default: ]
-
-nn,--n_neighbors<n_neighbors>¶ Number nearest neighbors. [default: 8]
plot_image¶
Plots the whole slide image supplied.
pathflowai-visualize plot_image [OPTIONS]
Options
-
-i,--image_file<image_file>¶ Input image file. [default: ./inputs/a.svs]
-
-cf,--compression_factor<compression_factor>¶ How much compress image. [default: 3.0]
-
-o,--outputfname<outputfname>¶ Output extracted image. [default: ./output_image.png]
plot_image_umap_embeddings¶
Plots a UMAP embedding with each point as its corresponding patch image.
pathflowai-visualize plot_image_umap_embeddings [OPTIONS]
Options
-
-i,--input_dir<input_dir>¶ Input directory for patches. [default: ./inputs/]
-
-e,--embeddings_file<embeddings_file>¶ Embeddings. [default: predictions/embeddings.pkl]
-
-b,--basename<basename>¶ Basename of patches. [default: ]
-
-o,--outputfilename<outputfilename>¶ Embedding visualization. [default: predictions/shap_plots.png]
-
-mpl,--mpl_scatter¶ Plot segmentations. [default: False]
-
-rb,--remove_background_annotation<remove_background_annotation>¶ If selected, removes 100% background patches based on this annotation. [default: ]
-
-ma,--max_background_area<max_background_area>¶ Max background area before exclusion. [default: 0.05]
-
-z,--zoom<zoom>¶ Size of images. [default: 0.05]
-
-nn,--n_neighbors<n_neighbors>¶ Number nearest neighbors. [default: 8]
-
-sc,--sort_col<sort_col>¶ Sort samples on this column. [default: ]
-
-sm,--sort_mode<sort_mode>¶ Sort ascending or descending. [default: asc]
- Options
asc|desc
plot_predictions¶
Overlays classification, regression and segmentation patch level predictions on top of whole slide image.
pathflowai-visualize plot_predictions [OPTIONS]
Options
-
-i,--input_dir<input_dir>¶ Input directory for patches. [default: ./inputs/]
-
-b,--basename<basename>¶ Basename of patches. [default: A01]
-
-p,--patch_info_file<patch_info_file>¶ Datbase containing all patches [default: patch_info.db]
-
-ps,--patch_size<patch_size>¶ Patch size. [default: 224]
-
-o,--outputfname<outputfname>¶ Output extracted image. [default: ./output_image.png]
-
-an,--annotations¶ Plot annotations instead of predictions. [default: False]
-
-cf,--compression_factor<compression_factor>¶ How much compress image. [default: 3.0]
-
-al,--alpha<alpha>¶ How much to give annotations/predictions versus original image. [default: 0.8]
-
-s,--segmentation¶ Plot segmentations. [default: False]
-
-sc,--n_segmentation_classes<n_segmentation_classes>¶ Number segmentation classes [default: 4]
-
-c,--custom_segmentation<custom_segmentation>¶ Add custom segmentation map from prediction, npy format. [default: ]
-
-ac,--annotation_col<annotation_col>¶ Column of annotations [default: annotation]
-
-sf,--scaling_factor<scaling_factor>¶ Multiply all prediction scores by this amount. [default: 1.0]
-
-tif,--tif_file¶ Write to tiff file. [default: False]
shapley_plot¶
Run SHAPley attribution method on patches after classification task to see where model made prediction based on.
pathflowai-visualize shapley_plot [OPTIONS]
Options
-
-m,--model_pkl<model_pkl>¶ Plotly output file. [default: ]
-
-bs,--batch_size<batch_size>¶ Batch size. [default: 32]
-
-o,--outputfilename<outputfilename>¶ SHAPley visualization. [default: predictions/shap_plots.png]
-
-mth,--method<method>¶ Method of explaining. [default: deep]
- Options
deep|gradient
-
-l,--local_smoothing<local_smoothing>¶ Local smoothing of SHAP scores. [default: 0.0]
-
-ns,--n_samples<n_samples>¶ Number shapley samples for shapley regression (gradient explainer). [default: 32]
-
-p,--pred_out<pred_out>¶ If not none, output prediction as shap label. [default: none]
- Options
none|sigmoid|softmax
pathflowai-monitor¶
pathflowai-monitor [OPTIONS] COMMAND [ARGS]...
Options
-
--version¶ Show the version and exit.
monitor_usage¶
Monitor Usage over Time Interval.
pathflowai-monitor monitor_usage [OPTIONS]
Options
-
-csv,--records_output_csv<records_output_csv>¶ Where to store records. [default: records.csv]
-
-tt,--total_time<total_time>¶ Total time to monitor for in minutes. [default: 1.0]
-
-dt,--delay_time<delay_time>¶ Time between samples, in seconds. [default: 1.0]
datasets.py¶
Houses the DynamicImageDataset class, also functions to help with image color channel normalization, transformers, etc..
-
class
pathflowai.datasets.DynamicImageDataset(dataset_df, set, patch_info_file, transformers, input_dir, target_names, pos_annotation_class, other_annotations=[], segmentation=False, patch_size=224, fix_names=True, target_segmentation_class=-1, target_threshold=0.0, oversampling_factor=1.0, n_segmentation_classes=4, gdl=False, mt_bce=False, classify_annotations=False)[source]¶ Generate image dataset that accesses images and annotations via dask.
- Parameters
- dataset_df:dataframe
Dataframe with WSI, which set it is in (train/test/val) and corresponding WSI labels if applicable.
- set:str
Whether train, test, val or pass (normalization) set.
- patch_info_file:str
SQL db with positional and annotation information on each slide.
- transformers:dict
Contains transformers to apply on images.
- input_dir:str
Directory where images comes from.
- target_names:list/str
Names of initial targets, which may be modified.
- pos_annotation_class:str
If selected and predicting on WSI, this class is labeled as a positive from the WSI, while the other classes are not.
- other_annotations:list
Other annotations to consider from patch info db.
- segmentation:bool
Conducting segmentation task?
- patch_size:int
Patch size.
- fix_names:bool
Whether to change the names of dataset_df.
- target_segmentation_class:list
Now can be used for classification as well, matched with two below options, samples images only from this class. Can specify this and below two options multiple times.
- target_threshold:list
Sampled only if above this threshold of occurence in the patches.
- oversampling_factor:list
Over sample them at this amount.
- n_segmentation_classes:int
Number classes to segment.
- gdl:bool
Using generalized dice loss?
- mt_bce:bool
For multi-target prediction tasks.
- classify_annotations:bool
For classifying annotations.
Methods
binarize_annotations(self[, binarizer, …])Label binarize some annotations or threshold them if classifying slide annotations.
concat(self, other_dataset)Concatenate this dataset with others.
get_class_weights(self[, i])Weight loss function with weights inversely proportional to the class appearence.
retain_ID(self, ID)Reduce the sample set to just images from one ID.
split_by_ID(self)Generator similar to groupby, but splits up by ID, generates (ID,data) using retain_ID.
subsample(self, p)Sample subset of dataset.
-
binarize_annotations(self, binarizer=None, num_targets=1, binary_threshold=0.0)[source]¶ Label binarize some annotations or threshold them if classifying slide annotations.
- Parameters
- binarizer:LabelBinarizer
Binarizes the labels of a column(s)
- num_targets:int
Number of desired targets to preidict on.
- binary_threshold:float
Amount of annotation in patch before positive annotation.
- Returns
- binarizer
-
concat(self, other_dataset)[source]¶ Concatenate this dataset with others. Updates its own internal attributes.
- Parameters
- other_dataset:DynamicImageDataset
Other image dataset.
-
get_class_weights(self, i=0)[source]¶ Weight loss function with weights inversely proportional to the class appearence.
- Parameters
- i:int
If multi-target, class used for weighting.
- Returns
- self
Dataset.
-
retain_ID(self, ID)[source]¶ Reduce the sample set to just images from one ID.
- Parameters
- ID:str
Basename/ID to predict on.
- Returns
- self
-
pathflowai.datasets.RandomRotate90()[source]¶ Transformer for random 90 degree rotation image.
- Returns
- function
Transformer function for operation.
-
pathflowai.datasets.create_transforms(mean, std)[source]¶ Create transformers.
- Parameters
- mean:list
See get_data_transforms.
- std:list
See get_data_transforms.
- Returns
- dict
Transformers.
-
pathflowai.datasets.get_data_transforms(patch_size=None, mean=[], std=[], resize=False, transform_platform='torch', elastic=True)[source]¶ Get data transformers for training test and validation sets.
- Parameters
- patch_size:int
Original patch size being transformed.
- mean:list of float
Mean RGB
- std:list of float
Std RGB
- resize:int
Which patch size to resize to.
- transform_platform:str
Use pytorch or albumentation transforms.
- elastic:bool
Whether to add elastic deformations from albumentations.
- Returns
- dict
Transformers.
-
pathflowai.datasets.get_normalizer(normalization_file, dataset_opts)[source]¶ Find mean and standard deviation of images in batches.
- Parameters
- normalization_file:str
File to store normalization information.
- dataset_opts:type
Dictionary storing information to create DynamicDataset class.
- Returns
- dict
Stores RGB mean, stdev.
losses.py¶
Some additional loss functions that can be called using the pipeline, some of which still to be implemented.
-
class
pathflowai.losses.FocalLoss(num_class, alpha=None, gamma=2, balance_index=-1, smooth=None, size_average=True)[source]¶ # https://raw.githubusercontent.com/Hsuxu/Loss_ToolBox-PyTorch/master/FocalLoss/FocalLoss.py This is a implementation of Focal Loss with smooth label cross entropy supported which is proposed in ‘Focal Loss for Dense Object Detection. (https://arxiv.org/abs/1708.02002)’
Focal_Loss= -1*alpha*(1-pt)*log(pt)
- Parameters
num_class –
alpha – (tensor) 3D or 4D the scalar factor for this criterion
gamma – (float,double) gamma > 0 reduces the relative loss for well-classified examples (p>0.5) putting more focus on hard misclassified example
smooth – (float,double) smooth value when cross entropy
balance_index – (int) balance class index, should be specific when alpha is float
size_average – (bool, optional) By default, the losses are averaged over each loss element in the batch.
Methods
__call__(self, \*input, \*\*kwargs)Call self as a function.
add_module(self, name, module)Adds a child module to the current module.
apply(self, fn)Applies
fnrecursively to every submodule (as returned by.children()) as well as self.buffers(self[, recurse])Returns an iterator over module buffers.
children(self)Returns an iterator over immediate children modules.
cpu(self)Moves all model parameters and buffers to the CPU.
cuda(self[, device])Moves all model parameters and buffers to the GPU.
double(self)Casts all floating point parameters and buffers to
doubledatatype.eval(self)Sets the module in evaluation mode.
extra_repr(self)Set the extra representation of the module
float(self)Casts all floating point parameters and buffers to float datatype.
forward(self, logit, target)Defines the computation performed at every call.
half(self)Casts all floating point parameters and buffers to
halfdatatype.load_state_dict(self, state_dict[, strict])Copies parameters and buffers from
state_dictinto this module and its descendants.modules(self)Returns an iterator over all modules in the network.
named_buffers(self[, prefix, recurse])Returns an iterator over module buffers, yielding both the name of the buffer as well as the buffer itself.
named_children(self)Returns an iterator over immediate children modules, yielding both the name of the module as well as the module itself.
named_modules(self[, memo, prefix])Returns an iterator over all modules in the network, yielding both the name of the module as well as the module itself.
named_parameters(self[, prefix, recurse])Returns an iterator over module parameters, yielding both the name of the parameter as well as the parameter itself.
parameters(self[, recurse])Returns an iterator over module parameters.
register_backward_hook(self, hook)Registers a backward hook on the module.
register_buffer(self, name, tensor)Adds a persistent buffer to the module.
register_forward_hook(self, hook)Registers a forward hook on the module.
register_forward_pre_hook(self, hook)Registers a forward pre-hook on the module.
register_parameter(self, name, param)Adds a parameter to the module.
state_dict(self[, destination, prefix, …])Returns a dictionary containing a whole state of the module.
to(self, \*args, \*\*kwargs)Moves and/or casts the parameters and buffers.
train(self[, mode])Sets the module in training mode.
type(self, dst_type)Casts all parameters and buffers to
dst_type.zero_grad(self)Sets gradients of all model parameters to zero.
share_memory
-
forward(self, logit, target)[source]¶ Defines the computation performed at every call.
Should be overridden by all subclasses.
Note
Although the recipe for forward pass needs to be defined within this function, one should call the
Moduleinstance afterwards instead of this since the former takes care of running the registered hooks while the latter silently ignores them.
-
class
pathflowai.losses.GeneralizedDice(**kwargs)[source]¶ https://raw.githubusercontent.com/LIVIAETS/surface-loss/master/losses.py
Methods
__call__(self, probs, target, _)Call self as a function.
-
class
pathflowai.losses.GeneralizedDiceLoss(weight=None, channelwise=False, eps=1e-06, add_softmax=False)[source]¶ https://raw.githubusercontent.com/inferno-pytorch/inferno/0561e8a95cde6bfc5e10a3609841b7b0ca5b03ca/inferno/extensions/criteria/set_similarity_measures.py Computes the scalar Generalized Dice Loss defined in https://arxiv.org/abs/1707.03237
This version works for multiple classes and expects predictions for every class (e.g. softmax output) and one-hot targets for every class.
Methods
__call__(self, \*input, \*\*kwargs)Call self as a function.
add_module(self, name, module)Adds a child module to the current module.
apply(self, fn)Applies
fnrecursively to every submodule (as returned by.children()) as well as self.buffers(self[, recurse])Returns an iterator over module buffers.
children(self)Returns an iterator over immediate children modules.
cpu(self)Moves all model parameters and buffers to the CPU.
cuda(self[, device])Moves all model parameters and buffers to the GPU.
double(self)Casts all floating point parameters and buffers to
doubledatatype.eval(self)Sets the module in evaluation mode.
extra_repr(self)Set the extra representation of the module
float(self)Casts all floating point parameters and buffers to float datatype.
forward(self, input, target)input: torch.FloatTensor or torch.cuda.FloatTensor target: torch.FloatTensor or torch.cuda.FloatTensor
half(self)Casts all floating point parameters and buffers to
halfdatatype.load_state_dict(self, state_dict[, strict])Copies parameters and buffers from
state_dictinto this module and its descendants.modules(self)Returns an iterator over all modules in the network.
named_buffers(self[, prefix, recurse])Returns an iterator over module buffers, yielding both the name of the buffer as well as the buffer itself.
named_children(self)Returns an iterator over immediate children modules, yielding both the name of the module as well as the module itself.
named_modules(self[, memo, prefix])Returns an iterator over all modules in the network, yielding both the name of the module as well as the module itself.
named_parameters(self[, prefix, recurse])Returns an iterator over module parameters, yielding both the name of the parameter as well as the parameter itself.
parameters(self[, recurse])Returns an iterator over module parameters.
register_backward_hook(self, hook)Registers a backward hook on the module.
register_buffer(self, name, tensor)Adds a persistent buffer to the module.
register_forward_hook(self, hook)Registers a forward hook on the module.
register_forward_pre_hook(self, hook)Registers a forward pre-hook on the module.
register_parameter(self, name, param)Adds a parameter to the module.
state_dict(self[, destination, prefix, …])Returns a dictionary containing a whole state of the module.
to(self, \*args, \*\*kwargs)Moves and/or casts the parameters and buffers.
train(self[, mode])Sets the module in training mode.
type(self, dst_type)Casts all parameters and buffers to
dst_type.zero_grad(self)Sets gradients of all model parameters to zero.
share_memory
-
class
pathflowai.losses.SurfaceLoss(**kwargs)[source]¶ https://raw.githubusercontent.com/LIVIAETS/surface-loss/master/losses.py
Methods
__call__(self, probs, dist_maps, _)Call self as a function.
-
pathflowai.losses.assert_(condition, message='', exception_type=<class 'AssertionError'>)[source]¶ https://raw.githubusercontent.com/inferno-pytorch/inferno/0561e8a95cde6bfc5e10a3609841b7b0ca5b03ca/inferno/utils/exceptions.py Like assert, but with arbitrary exception types.
-
pathflowai.losses.class2one_hot(seg:torch.Tensor, C:int) → torch.Tensor[source]¶ https://raw.githubusercontent.com/LIVIAETS/surface-loss/master/utils.py
-
pathflowai.losses.eq(a:torch.Tensor, b) → bool[source]¶ https://raw.githubusercontent.com/LIVIAETS/surface-loss/master/utils.py
-
pathflowai.losses.flatten_samples(input_)[source]¶ https://raw.githubusercontent.com/inferno-pytorch/inferno/0561e8a95cde6bfc5e10a3609841b7b0ca5b03ca/inferno/utils/torch_utils.py Flattens a tensor or a variable such that the channel axis is first and the sample axis is second. The shapes are transformed as follows:
(N, C, H, W) –> (C, N * H * W) (N, C, D, H, W) –> (C, N * D * H * W) (N, C) –> (C, N)
The input must be atleast 2d.
-
pathflowai.losses.one_hot(t:torch.Tensor, axis=1) → bool[source]¶ https://raw.githubusercontent.com/LIVIAETS/surface-loss/master/utils.py
-
pathflowai.losses.one_hot2dist(seg:numpy.ndarray) → numpy.ndarray[source]¶ https://raw.githubusercontent.com/LIVIAETS/surface-loss/master/utils.py
-
pathflowai.losses.simplex(t:torch.Tensor, axis=1) → bool[source]¶ https://raw.githubusercontent.com/LIVIAETS/surface-loss/master/utils.py
-
pathflowai.losses.sset(a:torch.Tensor, sub:Iterable) → bool[source]¶ https://raw.githubusercontent.com/LIVIAETS/surface-loss/master/utils.py
-
pathflowai.losses.uniq(a:torch.Tensor) → Set[source]¶ https://raw.githubusercontent.com/LIVIAETS/surface-loss/master/utils.py
sampler.py¶
Balanced sampling based on one of the columns of the patch information.
-
class
pathflowai.sampler.ImbalancedDatasetSampler(dataset, indices=None, num_samples=None)[source]¶ Samples elements randomly from a given list of indices for imbalanced dataset https://raw.githubusercontent.com/ufoym/imbalanced-dataset-sampler/master/sampler.py Arguments:
indices (list, optional): a list of indices num_samples (int, optional): number of samples to draw
schedulers.py¶
Modulates the learning rate during the training process.
-
class
pathflowai.schedulers.CosineAnnealingWithRestartsLR(optimizer, T_max, eta_min=0, last_epoch=-1, T_mult=1.0, alpha_decay=1.0)[source]¶ Set the learning rate of each parameter group using a cosine annealing schedule, where \(\eta_{max}\) is set to the initial lr and \(T_{cur}\) is the number of epochs since the last restart in SGDR:
\[\eta_t = \eta_{min} + \frac{1}{2}(\eta_{max} - \eta_{min})(1 + \cos(\frac{T_{cur}}{T_{max}}\pi))\]When last_epoch=-1, sets initial lr as lr. It has been proposed in
SGDR: Stochastic Gradient Descent with Warm Restarts. This implements the cosine annealing part of SGDR, the restarts and number of iterations multiplier.
- Args:
optimizer (Optimizer): Wrapped optimizer. T_max (int): Maximum number of iterations. T_mult (float): Multiply T_max by this number after each restart. Default: 1. eta_min (float): Minimum learning rate. Default: 0. last_epoch (int): The index of last epoch. Default: -1.
- Attributes
- step_n
Methods
load_state_dict(self, state_dict)Loads the schedulers state.
state_dict(self)Returns the state of the scheduler as a
dict.cosine
get_lr
restart
step
-
class
pathflowai.schedulers.Scheduler(optimizer=None, opts={'T_max': 10, 'T_mult': 2, 'eta_min': 5e-08, 'lr_scheduler_decay': 0.5, 'scheduler': 'null'})[source]¶ Scheduler class that modulates learning rate of torch optimizers over epochs.
- Parameters
- optimizertype
torch.Optimizer object
- optstype
Options of setting the learning rate scheduler, see default.
- Attributes
- schedulerstype
Different types of schedulers to choose from.
- scheduler_step_fntype
How scheduler updates learning rate.
- initial_lrtype
Initial set learning rate.
- scheduler_choicetype
What scheduler type was chosen.
- schedulertype
Scheduler object chosen that will more directly update optimizer LR.
Methods
get_lr(self)Return current learning rate.
step(self)Update optimizer learning rate
visualize.py¶
Plots SHAP outputs, UMAP embeddings, and overlays predictions on top of WSI.
-
class
pathflowai.visualize.PlotlyPlot[source]¶ Creates plotly html plots.
Methods
add_plot(self, t_data_df[, G, color_col, …])Adds plotting data to be plotted.
plot(self, output_fname[, axes_off])Plot embedding of patches to html file.
-
add_plot(self, t_data_df, G=None, color_col='color', name_col='name', xyz_cols=['x', 'y', 'z'], size=2, opacity=1.0, custom_colors=[])[source]¶ Adds plotting data to be plotted.
- Parameters
- t_data_df:dataframe
3-D transformed dataframe.
- G:nx.Graph
Networkx graph.
- color_col:str
Column to use to color points.
- name_col:str
Column to use to name points.
- xyz_cols:list
3 columns that denote x,y,z coords.
- size:int
Marker size.
- opacity:float
Marker opacity.
- custom_colors:list
Custom colors to supply.
-
-
class
pathflowai.visualize.PredictionPlotter(dask_arr_dict, patch_info_db, compression_factor=3, alpha=0.5, patch_size=224, no_db=False, plot_annotation=False, segmentation=False, n_segmentation_classes=4, input_dir='', annotation_col='annotation', scaling_factor=1.0)[source]¶ Plots predictions over entire image.
- Parameters
- dask_arr_dict:dict
Stores all dask arrays corresponding to all of the images.
- patch_info_db:str
Patch level information, eg. prediction.
- compression_factor:float
How much to compress image by.
- alpha:float
Low value assigns higher weight to prediction over original image.
- patch_size:int
Patch size.
- no_db:bool
Don’t use patch information.
- plot_annotation:bool
Plot annotations from patch information.
- segmentation:bool
Plot segmentation mask.
- n_segmentation_classes:int
Number segmentation classes.
- input_dir:str
Input directory.
- annotation_col:str
Annotation column to plot.
- scaling_factor:float
Multiplies the prediction scores to make them appear darker on the images when predicting.
Methods
add_custom_segmentation(self, basename, npy)Replace segmentation mask with new custom segmentation.
generate_image(self, ID)Generate the image array for the whole slide image with predictions overlaid.
output_image(self, img, filename[, tif])Output calculated image to file.
return_patch(self, ID, x, y, patch_size)Return one single patch instead of entire image.
-
add_custom_segmentation(self, basename, npy)[source]¶ Replace segmentation mask with new custom segmentation.
- Parameters
- basename:str
Patient ID
- npy:str
Numpy mask.
-
generate_image(self, ID)[source]¶ Generate the image array for the whole slide image with predictions overlaid.
- Parameters
- ID:str
patient ID.
- Returns
- array
Resulting overlaid whole slide image.
-
pathflowai.visualize.annotation2rgb(i, palette, arr)[source]¶ Go from annotation of patch to color.
- Parameters
- i:int
Annotation index.
- palette:palette
Index to color mapping.
- arr:array
Image array.
- Returns
- array
Resulting image.
-
pathflowai.visualize.blend(arr1, arr2, alpha=0.5)[source]¶ Blend 2 arrays together, mixing with alpha.
- Parameters
- arr1:array
Image 1.
- arr2:array
Image 2.
- alpha:float
Higher alpha makes image more like image 1.
- Returns
- array
Resulting image.
-
pathflowai.visualize.plot_image_(image_file, compression_factor=2.0, test_image_name='test.png')[source]¶ Plots entire SVS/other image.
- Parameters
- image_file:str
Image file.
- compression_factor:float
Amount to shrink each dimension of image.
- test_image_name:str
Output image file.
-
pathflowai.visualize.plot_shap(model, dataset_opts, transform_opts, batch_size, outputfilename, n_outputs=1, method='deep', local_smoothing=0.0, n_samples=20, pred_out=False)[source]¶ Plot shapley attributions overlaid on images for classification tasks.
- Parameters
- model:nn.Module
Pytorch model.
- dataset_opts:dict
Options used to configure dataset
- transform_opts:dict
Options used to configure transformers.
- batch_size:int
Batch size for training.
- outputfilename:str
Output filename.
- n_outputs:int
Number of top outputs.
- method:str
Gradient or deep explainer.
- local_smoothing:float
How much to smooth shapley map.
- n_samples:int
Number shapley samples to draw.
- pred_out:bool
Label images with binary prediction score?
-
pathflowai.visualize.plot_umap_images(dask_arr_dict, embeddings_file, ID=None, cval=1.0, image_res=300.0, outputfname='output_embedding.png', mpl_scatter=True, remove_background_annotation='', max_background_area=0.01, zoom=0.05, n_neighbors=10, sort_col='', sort_mode='asc')[source]¶ Make UMAP embedding plot, overlaid with images.
- Parameters
- dask_arr_dict:dict
Stored dask arrays for each WSI.
- embeddings_file:str
Embeddings pickle file stored from running using after trainign the model.
- ID:str
Patient ID.
- cval:float
Deprecated
- image_res:float
Image resolution.
- outputfname:str
Output image file.
- mpl_scatter:bool
Recommended: Use matplotlib for scatter plot.
- remove_background_annotation:str
Remove the background annotations. Enter for annotation to remove.
- max_background_area:float
Maximum backgrund area in each tile for inclusion.
- zoom:float
How much to zoom in on each patch, less than 1 is zoom out.
- n_neighbors:int
Number of neighbors for UMAP embedding.
- sort_col:str
Patch info column to sort on.
- sort_mode:str
Sort ascending or descending.
- Returns
- type
Description of returned object.
- Inspired by: https://gist.github.com/lukemetz/be6123c7ee3b366e333a
- WIP!! Needs testing.
-
pathflowai.visualize.prob2rbg(prob, palette, arr)[source]¶ Convert probability score to rgb image.
- Parameters
- prob:float
Between 0 and 1 score.
- palette:palette
Pallet converts between prob and color.
- arr:array
Original array.
- Returns
- array
New image colored by prediction score.
utils.py¶
General utilities that still need to be broken up into preprocessing, machine learning input preparation, and output submodules.
-
pathflowai.utils.add_purple_mask(arr)[source]¶ Optional add intensity mask to the dask array.
- Parameters
- arr:dask.array
Image data.
- Returns
- array
Image data with intensity added as forth channel.
-
pathflowai.utils.adjust_mask(mask_file, dask_img_array_file, out_npy, n_neighbors)[source]¶ Fixes segmentation masks to reduce coarse annotations over empty regions.
- Parameters
- mask_file:str
NPY segmentation mask.
- dask_img_array_file:str
Dask image file.
- out_npy:str
Output numpy file.
- n_neighbors:int
Number nearest neighbors for dilation and erosion of mask from background to not background.
- Returns
- str
Output numpy file.
-
pathflowai.utils.create_purple_mask(arr, img_size=None, sparse=True)[source]¶ Create a gray scale intensity mask. This will be changed soon to support other thresholding QC methods.
- Parameters
- arr:dask.array
Dask array containing image information.
- img_size:int
Deprecated.
- sparse:bool
Deprecated
- Returns
- dask.array
Intensity, grayscale array over image.
-
pathflowai.utils.create_sparse_annotation_arrays(xml_file, img_size, annotations=[])[source]¶ Convert annotation xml to shapely objects and store in dictionary.
- Parameters
- xml_file:str
XML file containing annotations.
- img_size:int
Deprecated.
- annotations:list
Annotations to look for in xml export.
- Returns
- dict
Dictionary with annotation-shapely object pairs.
-
pathflowai.utils.create_train_val_test(train_val_test_pkl, input_info_db, patch_size)[source]¶ Create dataframe that splits slides into training validation and test.
- Parameters
- train_val_test_pkl:str
Pickle for training validation and test slides.
- input_info_db:str
Patch information SQL database.
- patch_size:int
Patch size looking to access.
- Returns
- dataframe
Train test validation splits.
-
pathflowai.utils.df2sql(df, sql_file, patch_size, mode='replace')[source]¶ Write dataframe containing patch level information to SQL db.
- Parameters
- df:dataframe
Dataframe containing patch information.
- sql_file:str
SQL database.
- patch_size:int
Size of patches.
- mode:str
Replace or append.
-
pathflowai.utils.extract_patch_information(basename, input_dir='./', annotations=[], threshold=0.5, patch_size=224, generate_finetune_segmentation=False, target_class=0, intensity_threshold=100.0, target_threshold=0.0, adj_mask='', basic_preprocess=False, tries=0)[source]¶ Final step of preprocessing pipeline. Break up image into patches, include if not background and of a certain intensity, find area of each annotation type in patch, spatial information, image ID and dump data to SQL table.
- Parameters
- basename:str
Patient ID.
- input_dir:str
Input directory.
- annotations:list
List of annotations to record, these can be different tissue types, must correspond with XML labels.
- threshold:float
Value between 0 and 1 that indicates the minimum amount of patch that musn’t be background for inclusion.
- patch_size:int
Patch size of patches; this will become one of the tables.
- generate_finetune_segmentation:bool
Deprecated.
- target_class:int
Number of segmentation classes desired, from 0th class to target_class-1 will be annotated in SQL.
- intensity_threshold:float
Value between 0 and 255 that represents minimum intensity to not include as background. Will be modified with new transforms.
- target_threshold:float
Deprecated.
- adj_mask:str
Adjusted mask if performed binary opening operations in previous preprocessing step.
- basic_preprocess:bool
Do not store patch level information.
- tries:int
Number of tries in case there is a Dask timeout, run again.
- Returns
- dataframe
Patch information.
-
pathflowai.utils.generate_patch_pipeline(basename, input_dir='./', annotations=[], threshold=0.5, patch_size=224, out_db='patch_info.db', generate_finetune_segmentation=False, target_class=0, intensity_threshold=100.0, target_threshold=0.0, adj_mask='', basic_preprocess=False)[source]¶ Short summary.
- Parameters
- basename:str
Patient ID.
- input_dir:str
Input directory.
- annotations:list
List of annotations to record, these can be different tissue types, must correspond with XML labels.
- threshold:float
Value between 0 and 1 that indicates the minimum amount of patch that musn’t be background for inclusion.
- patch_size:int
Patch size of patches; this will become one of the tables.
- out_db:str
Output SQL database.
- generate_finetune_segmentation:bool
Deprecated.
- target_class:int
Number of segmentation classes desired, from 0th class to target_class-1 will be annotated in SQL.
- intensity_threshold:float
Value between 0 and 255 that represents minimum intensity to not include as background. Will be modified with new transforms.
- target_threshold:float
Deprecated.
- adj_mask:str
Adjusted mask if performed binary opening operations in previous preprocessing step.
- basic_preprocess:bool
Do not store patch level information.
-
pathflowai.utils.img2npy_(input_dir, basename, svs_file)[source]¶ Convert SVS, TIF, TIFF to NPY.
- Parameters
- input_dir:str
Output file dir.
- basename:str
Basename of output file
- svs_file:str
SVS, TIF, TIFF file input.
- Returns
- str
NPY output file.
-
pathflowai.utils.is_coords_in_box(coords, patch_size, boxes)[source]¶ Get area of annotation in patch.
- Parameters
- coords:array
X,Y coordinates of patch.
- patch_size:int
Patch size.
- boxes:list
Shapely objects for annotations.
- Returns
- float
Area of annotation type.
-
pathflowai.utils.is_image_in_boxes(image_coord_dict, boxes)[source]¶ Find if image intersects with annotations.
- Parameters
- image_coord_dict:dict
Dictionary of patches.
- boxes:list
Shapely annotation shapes.
- Returns
- dict
Dictionary of whether image intersects with any of the annotations.
-
pathflowai.utils.is_valid_patch(xs, ys, patch_size, purple_mask, intensity_threshold, threshold=0.5)[source]¶ Deprecated, computes whether patch is valid.
-
pathflowai.utils.load_dataset(in_zarr, in_pkl)[source]¶ Load ZARR image and annotations pickle.
- Parameters
- in_zarr:str
Input image.
- in_pkl:str
Input annotations.
- Returns
- dask.array
Image array.
- dict
Annotations dictionary.
-
pathflowai.utils.load_image(svs_file)[source]¶ Load SVS, TIF, TIFF
- Parameters
- svs_file:type
Description of parameter svs_file.
- Returns
- type
Description of returned object.
-
pathflowai.utils.load_process_image(svs_file, xml_file=None, npy_mask=None, annotations=[])[source]¶ Load SVS-like image (including NPY), segmentation/classification annotations, generate dask array and dictionary of annotations.
- Parameters
- svs_file:str
Image file
- xml_file:str
Annotation file.
- npy_mask:array
Numpy segmentation mask.
- annotations:list
List of annotations in xml.
- Returns
- array
Dask array of image.
- dict
Annotation masks.
-
pathflowai.utils.load_sql_df(sql_file, patch_size)[source]¶ Load pandas dataframe from SQL, accessing particular patch size within SQL.
- Parameters
- sql_file:str
SQL db.
- patch_size:int
Patch size.
- Returns
- dataframe
Patch level information.
-
modify_patch_info(input_info_db='patch_info.db', slide_labels=Empty DataFrame -
Columns: [] -
Index: [], pos_annotation_class='', patch_size=224, segmentation=False, other_annotations=[], target_segmentation_class=-1, target_threshold=0.0, classify_annotations=False) Modify the patch information to get ready for deep learning, incorporate whole slide labels if needed.
- Parameters
- input_info_db:str
SQL DB file.
- slide_labels:dataframe
Dataframe with whole slide labels.
- pos_annotation_class:str
Tissue/annotation label to label with whole slide image label, if not supplied, any slide’s patches receive the whole slide label.
- patch_size:int
Patch size.
- segmentation:bool
Segmentation?
- other_annotations:list
Other annotations to access from patch information.
- target_segmentation_class:int
Segmentation class to threshold.
- target_threshold:float
Include patch if patch has target area greater than this.
- classify_annotations:bool
Classifying annotations for pretraining, or final model?
- Returns
- dataframe
Modified patch information.
-
pathflowai.utils.npy2da(npy_file)[source]¶ Numpy to dask array.
- Parameters
- npy_file:str
Input npy file.
- Returns
- dask.array
Converted numpy array to dask.
-
pathflowai.utils.parse_coord_return_boxes(xml_file, annotation_name='', return_coords=False)[source]¶ Get list of shapely objects for each annotation in the XML object.
- Parameters
- xml_file:str
Annotation file.
- annotation_name:str
Name of xml annotation.
- return_coords:bool
Just return list of coords over shapes.
- Returns
- list
List of shapely objects.
-
pathflowai.utils.process_svs(svs_file, xml_file, annotations=[], output_dir='./')[source]¶ Store images into npy format and store annotations into pickle dictionary.
- Parameters
- svs_file:str
Image file.
- xml_file:str
Annotations file.
- annotations:list
List of annotations in image.
- output_dir:str
Output directory.
-
pathflowai.utils.return_image_coord(nx=0, ny=0, xl=3333, yl=3333, xi=0, yi=0, xc=3, yc=3, dimx=224, dimy=224, output_point=False)[source]¶ Deprecated
-
pathflowai.utils.run_preprocessing_pipeline(svs_file, xml_file=None, npy_mask=None, annotations=[], out_zarr='output_zarr.zarr', out_pkl='output.pkl')[source]¶ Run preprocessing pipeline. Store image into zarr format, segmentations maintain as npy, and xml annotations as pickle.
- Parameters
- svs_file:str
Input image file.
- xml_file:str
Input annotation file.
- npy_mask:str
NPY segmentation mask.
- annotations:list
List of annotations.
- out_zarr:str
Output zarr for image.
- out_pkl:str
Output pickle for annotations.
-
pathflowai.utils.save_all_patch_info(basenames, input_dir='./', annotations=[], threshold=0.5, patch_size=224, output_pkl='patch_info.pkl')[source]¶ Deprecated.
-
pathflowai.utils.save_dataset(arr, masks, out_zarr, out_pkl)[source]¶ Saves dask array image, dictionary of annotations to zarr and pickle respectively.
- Parameters
- arr:array
Image.
- masks:dict
Dictionary of annotation shapes.
- out_zarr:str
Zarr output file for image.
- out_pkl:str
Pickle output file.
-
pathflowai.utils.segmentation_predictions2npy(y_pred, patch_info, segmentation_map, npy_output)[source]¶ Convert segmentation predictions from model to numpy masks.
- Parameters
- y_pred:list
List of patch segmentation masks
- patch_info:dataframe
Patch information from DB.
- segmentation_map:array
Existing segmentation mask.
- npy_output:str
Output npy file.
-
pathflowai.utils.svs2dask_array(svs_file, tile_size=1000, overlap=0, remove_last=True, allow_unknown_chunksizes=False)[source]¶ Convert SVS, TIF or TIFF to dask array.
- Parameters
- svs_file:str
Image file.
- tile_size:int
Size of chunk to be read in.
- overlap:int
Do not modify, overlap between neighboring tiles.
- remove_last:bool
Remove last tile because it has a custom size.
- allow_unknown_chunksizes: bool
Allow different chunk sizes, more flexible, but slowdown.
- Returns
- dask.array
Dask Array.
>>> arr=svs2dask_array(svs_file, tile_size=1000, overlap=0, remove_last=True, allow_unknown_chunksizes=False) ..
>>> arr2=arr.compute() ..
>>> arr3=to_pil(cv2.resize(arr2, dsize=(1440,700), interpolation=cv2.INTER_CUBIC)) ..
>>> arr3.save(test_image_name) ..